Replication and Reproduction
The one property that distinguishes biological processes from most other physical processes is that life is able to reproduce. How this occurs has been explained in several ways, either as an autocatalytic process or as an informational process, or both. Analogous issues arise in other domains, such as cultural evolution. The notion of replication has been proposed as the sine qua non of evolution, while reproduction has classically been a defining feature of life. Even more recent developments include a return to prominence of reproduction and its physical aspects over information-based accounts.
- 1. Historical Background
- 2. The Conceptual Grip of Genes and Organisms
- 3. Dawkins on Replicators and Vehicles
- 4. Replicators and Genes
- 5. Memes and the Immune System
- 6. The Extended Replicator
- 7. Information
- 8. Reproducers
- 9. Conclusion
- Bibliography
- Academic Tools
- Other Internet Resources
- Related Entries
1. Historical Background
The basic idea of replication has a long history in biology. Initially it arose as a concern in the period immediately after the second world war, with the rise of information theory, communication theory, cybernetics, and Erwin Schrödinger's book What is Life? Biologists immediately began to consider the informational aspect of life, in each of these theoretical frameworks, in areas ranging from neurology to molecular biology, but especially the latter. Informational accounts of inheritance begin in the middle of the nineteenth century (Lewes 1856), but it was only after Schrödinger's book on the nature of life (1969 [1944]) and then the development of communications theory by Shannon (1948) and cybernetics by Wiener (1948) that inheritance in biology began to take on a specifically informational tint. The Central Dogma was initially cast in terms of structural information (Šustar 2007), and only later recast by Crick as a more cybernetic or computational kind of information (Crick 1970). In the late 1950s, after the structure of DNA had been widely understood, Jacobson proposed a cybernetic model of reproduction (1958), going so far as to make a literal model out of train sets and electronic circuits. Later, von Neumann proposed a theory of self-reproducing automata (von Neumann and Burks 1966) which has become a parallel but influential research program to the issue in biology (Mitchell 2009). Jacob (1973) cemented the logical description of heredity, but the ultimate source of the notion that genes are information comes from Williams' classic 1966 book Adaptation and Natural Selection, in which he referred to genes as “cybernetic abstractions” (p. 33). This book was one inspiration for Dawkins' Selfish Gene (1976), and much of what Dawkins proposed about replicators is an extension of Williams' views, in conjunction with those of Hamilton (1964) and various ethological ideas.
An early example of this is the distinction between homocatalysis and heterocatalysis and later between transcription and translation (Hodge 1989). The basic distinction that these pairs of terms were designed to indicate is between like producing like (homocatalysis and transcription) and like producing unlike (heterocatalysis and translation; since then, a different contrast has also come to the fore in the realization that heredity with mutations also means that heredity is like producing unlike (mutants) as well as offspring like parents, so that heredity should be glossed as like variants producing like variants, which cuts across the homo-/hetero-catalytic distinction.). The paradigm example of this distinction is the contrast between genes and organisms. Genes perform two functions. They make other genes and, by means of the developmental process, they help produce organisms and their phenotypic traits (for a discussion of these two processes at the molecular level, see Rebek 1994 and Cook 1999).
As pervasive as this terminology has been, it engendered very little controversy until Richard Dawkins introduced the distinction between replicators and vehicles in his The Selfish Gene (1976). For his purposes Dawkins found the contrast between genes and organisms too restrictive. Everyone agrees that genes are replicators, but genes may not be the only replicators. Perhaps more inclusive entities than single genes might also function as replicators. At the very least, this possibility should not be defined out of existence. Hence, Dawkins adopted “replicator” as a more inclusive and general term than “gene.” He also introduced the term “vehicle” for those entities produced by replicators that help these replicators increase in numbers by interacting effectively with their environments. This distinction can be expressed in terms of either entities or processes. According to Dawkins replicators function in replication, while vehicles function in environmental interaction.
A long-standing dispute in evolutionary biology concerns the levels at which selection can occur (Lloyd 1994 [1988]; Brandon 1996; Keller 1999; Godfrey-Smith and Kerr 2002; Okasha 2004, 2005, 2006). As it turns out, there is no one process termed “selection.” Instead there are two processes—”replication” and “environmental interaction”. Some authors see this dispute as concerning the levels at which replication can take place. Dawkins argues that replication in biological evolution occurs exclusively at the level of the genetic material. Hence, he is a gene replicationist. Other authors take the levels of selection dispute to concern environmental interaction and insist that environmental interaction can take place at a variety of levels from single genes, cells and organisms to colonies, demes and possibly entire species. Organisms certainly interact with their environments in ways that bias the transmission of their genes, but then so do entities that are both less inclusive than entire organisms (e.g., sperm cells) and more inclusive (e.g., beehives).
The term “replication” refers first and foremost to copying. The nature of this process of replication and the objections to it are the main topics of this entry. Genes are self-replicating molecules. Some critics interpret this claim to be asserting that a strand of DNA placed on a glass slide might all on its own start replicating. Of course, no one has ever held such a nonsense view. Genes replicate themselves, but only with the aid of highly complicated molecular machinery. Too often, however, the importance of this machinery goes unnoticed. To be sure, when we make copies on a Xerox machine, we are interested in the texts, figures or just scrawls that appear on these sheets of paper. We are not interested in how the Xerox machine works, even if it does all the work.
Another issue arises when replication is analyzed in terms of information. Numerous mechanisms exist for passing on information from one replicator to another. Sometimes the material in which the information is encoded gets passed on; sometimes not. For example, in the commonest sort of transcription in biology, replication is accompanied by the transmission of a substantial amount of physical material. However, in other sorts of information transfer, little or no matter is passed on. In this case, replication via DNA is aberrant, not typical. Other critics object to any use of terms associated with human languages to characterize biological replication. Talk about the genetic code is at best heuristic, at worst totally inappropriate. However, the major problem with treating replication in biological contexts is that we do not have an analysis of “information” and related terms up to the job.
As mentioned earlier, genes have two functions. One is replication. The other is the production of organisms and their phenotypic traits. The connections between genes and organisms (development) are notoriously controversial. The vast majority of literature on the topic of replication, environmental interaction and selection concerns the complexities involved in describing development. Does it make sense to refer to the “gene for eye color” or the “gene for homosexuality”? However, the topic of this contribution is replication, not development. In replication, objects are copied, genes producing genes and, more generally, replicators producing replicators. Making sense of this topic is intimidating enough without introducing the relation between replication and environmental interaction. Unfortunately, replication cannot be explicated adequately without at least touching on development and its constituent difficulties. For example, if replication were simply the passing on of structure largely intact without any subsequent translation, we would not be tempted to employ terms such as “information” in connection with it.
2. The Conceptual Grip of Genes and Organisms
Those who offer a more general analysis of “selection” try to avoid the conventional way of viewing this process. For example, according to the Central Dogma of molecular biology, information can be transferred from nucleic acid to nucleic acid or from nucleic acid to proteins. It cannot be transferred from protein to protein or from proteins to nucleic acid (Crick 1970). This asymmetry results from nucleic acids exhibiting what might be termed “potential information” while proteins do not. Given a particular stretch of DNA, it can give rise to a variety of proteins, depending on various environmental factors. In a particular interaction, a particular protein will be produced. This protein will be only one of the numerous proteins possible given that stretch of DNA. In the translation of potential information into an actual protein, lots of information is lost. Hence, it is no longer available to be read back into the DNA that produced it.
But what if information flow occurs without the participation of genes? For example, from season to season a butterfly feeds on and lays her eggs on the same sort of nasty tasting host plant. In the process she transfers the nasty taste of the host plant to herself and to her progeny. Then her progeny does the same. This looks very much like information being transferred, but not via the genes. Granted, genes play several important roles in this process, but not in the transference of information. As another example, what if proteins served as templates for other proteins? We would have copying but nothing that might be termed a “genetic code” (Godfrey Smith 2000). The point of the preceding examples is to indicate how closely we are tied to traditional ways of viewing replication and translation. For example, just about everyone assumes that the development of organisms from fertilized zygote to mature adult is the only relation between replication and environmental interaction. Development is one possible relation between replicators and environmental interaction but not the only one. Freeing ourselves from the perspective of genes, organisms and development is easier said than done.
In this article fundamentally different perspectives are introduced and compared—so fundamental that they deserve to be termed “metaphysical.” Although the weight of evidence can be brought to bear on these alternative formulations, it is rarely decisive. Rather the deciding factor is how well these different perspectives organize this data. What impact on our understanding of biological phenomena does switching to the Gene's Eye view produce? Does extending the phenotype hurt or help? How about extending the notion of replication? Do these more general concepts facilitate the formation of a general analysis of replication that applies at the very least to gene-based biological evolution but also to selection in other contexts? Any answer to these questions entails a strange mix of science and philosophy.
Dawkins' analysis gave rise to the present-day controversy over replication and replicators. The first issue concerns disagreements over the proper definition of “replicator.” Dawkins argues that genes and only genes function as replicators in biology. He also introduces vehicles as the entities that aid genes in their quest to become more prevalent, but gradually he reduces the role of vehicles until it is all but nonexistent. We then proceed to evaluate the various criticisms of genes as replicators and Dawkins' extended phenotype. Can various elements of the immune system function as replicators? How about conceptual change? Can it be profitably viewed as a selection process? The idea of copying is essential to this discussion. We then turn to Developmental Systems Theory and the attempt to extend the replicator. Genes are replicators, but possibly they are not the only replicators. Finally, we discuss all too briefly the role of information in replication, and the notion of reproducers as a solution to these problems.
3. Dawkins on Replicators and Vehicles
For Dawkins, selection results from an interplay between replicators and vehicles. Replicators produce copies of themselves, but they also produce entities that interact with their environments in such a way that replication is differential. Some replicators are passed on more profusely than others. For Dawkins, genes are the sole replicators in biological evolution. They are important in a second sense as well. As fascinating as all the complex adaptations that have arisen through selection may be, the results of this process matter in selection only if they are reflected in the content of their respective replicators. Dawkins is frequently (and inaccurately) castigated for being a genetic determinist, as if he thought that genes are sufficient to produce phenotypic traits, but he is aware of the crucial role that an organism's environment, including other organisms, plays in selection. A gene all by itself never did anything.
In Dawkins' early writings, replicators and vehicles played different but complementary and equally important roles in selection, but as Dawkins honed his view of the evolutionary process, vehicles became less and less fundamental. Initially, Dawkins was content to dethrone the organism from its pride of place in biology. It is an important focus of environmental interaction, but other entities, both below and above the organismic level, can also function as vehicles. In later writings Dawkins goes even further and argues that phenotypic traits are what really matter in selection and that they can be treated independently of their being organized into vehicles. More than that, features such as spider webs should be viewed as part of a spider's phenotype. Hence, Dawkins chose as the title of his second book The Extended Phenotype (1982a).
From the start Dawkins acknowledged that entities at a variety of levels can function as vehicles. The issue for Dawkins is replication, and according to Dawkins, genes and only genes can function as replicators in biological evolution. How big these genetic units are depends on such things as the prevalence of sex, the frequency of crossover and the intensity of selection.
If there were sex but no crossing-over, each chromosome would be a replicator, and we should speak of adaptations as being for the good of the chromosome. If there is no sex we can, by the same token, treat the entire genome of an asexual organism as a replicator. But the organism itself is not a replicator. (Dawkins 1982a: 95)
Dawkins presents two reasons for organisms not being able to function as replicators. The first is the one he uses to delineate evolutionary genes. As in the case of long stretches of DNA, organisms are too easily and frequently broken up to be considered units of replication. A second reason why whole organisms cannot function as replicators is that they cannot pass on changes in their structure. Phenotypic change may result in phenotypic change (as in bad-tasting butterflies), but these changes do not find their way to the genetic material. Although epigenetic mechanisms have been shown to be passed on across generations (Jablonka and Raz 2009), Lamarckian inheritance as originally defined does not occur (see entry on inheritance systems). The amount of DNA that counts as a replicator certainly varies, but according to Dawkins, nothing more inclusive than the genetic material functions as replicators in biological evolution.
Dawkins never lost his fascination with vehicular adaptations, a fascination that his critics denigrate as Panglossian adaptationism. He fills his books with adaptationist scenarios, some more firmly supported by data than others, but from the perspective of the structure of evolutionary theory, replicators are much more important than vehicles. For example, Dawkins argues at some length that adaptations are always for the good of replicators, not vehicles (Lloyd 1992, see entry on units and levels of selection). Vehicles exhibit these adaptations, but ultimately all adaptations must be explicable in terms of changes in gene frequencies. Thus, it comes as no surprise when Dawkins (1994: 617) proclaims that he “coined the term ‘vehicle’ not to praise it but to bury it.” As prevalent as organisms might be, as determinate as the causal roles that they play in selection are, reference to them can and must be omitted from any perspicuous characterization of selection in the evolutionary process. Dawkins is far from a genetic determinist, but he is certainly a genetic reductionist. Whether reductionism itself is good or bad is a moot question (Van Regenmortel and Hull 2002).
4. Replicators and Genes
Although Dawkins finds it desirable to replace genes with replicators in his general characterization of the evolutionary process, he says very little about this more general notion in his The Selfish Gene. Instead he discusses the special case of genes as replicators. The primacy of the genetic perspective in the characterization of replicators is claimed by some (Griesemer 2000a) to be one of the chief weaknesses not only in Dawkins' discussions but also in the work of his successors. Hull and his colleagues claim to be providing a general notion of replication, adequate to handle all different sorts of replication, when too often we are simply reading peculiarities of genetic replication into our general analysis of replication.
According to Dawkins, replicators have three basic properties—longevity, fecundity and copying-fidelity. Longevity means longevity in the form of copies through descent (Dawkins 1982a: 84; Hull 1980), although the stability of tokens is included in the definition in The Selfish Gene (Dawkins 1976: 18, 1999: 17). No gene as a physical body lasts all that long. In mitosis, a gene loses half of its substance at each replication. What endures is not the entity itself but the information incorporated in its structure. It is this information that is copied with such high fidelity. Mutations do occur, but at very low frequencies. Even so, in some organisms mutation rates must be too high because mechanisms have evolved that search out and repair such errors.
The main source of variation in genes, however, is not mutation but crossover. Pairs of homologous chromosomes line up next to each other at meiosis, crossover and recombine. For stretches of DNA in which different alleles exist, the result can be a change in information. Quite obviously, the shorter the stretch of DNA involved, the less likely that crossover will occur and the message changed. Dawkins appeals to such dismantling of entities to argue against organisms functioning as replicators. In sexual organisms, organisms themselves are torn apart and repeatedly assembled each generation (Caporael 2003). If long stretches of DNA lack the necessary identity by descent to function as replicators, then sexual organisms certainly lack it. However, some other explanation has to be provided for asexual organisms because they pass on their overall structure largely unchanged from generation to generation. For example, R. A. Fisher, in his The Genetical Theory of Natural Selection (1930) considered the entire genetic complement of asexual organisms to be a single gene, and this view has been repeated from time to time since.
Dawkins bypasses classical Mendelian genes and even molecular genes to adopt G. C. Williams' conception of evolutionary genes as the fundamental units of natural selection (Williams 1966). Dawkins (1976: 30) defines evolutionary genes as
any portion of chromosomal material which potentially lasts for enough generations to serve as a unit of natural selection.
The limits of these genes need not be absolutely sharp. Nor must all genes be of the same length. The greater the selection pressure, the smaller the gene. At bottom, selection takes place between alternative alleles residing at the same locus.
As far as a gene is concerned, its alleles are its deadly rivals, but other genes are just a part of its environment, comparable to temperature, food, and predators, or companions. (Dawkins 1976: 40)
Alleles cannot cooperate with each other; only compete. That is where the “selfish” in “selfish gene” comes in.
According to Dawkins (1976: 95), the selfish gene is not just one physical bit of DNA. It is “all replicas of a particular bit of DNA, distributed throughout the world.” Hence, genes do not form classes of spatiotemporally unrelated individuals but trees. They must be replicas. But being a replica is not enough. The linear repetition of the “same gene” in the form of several hundred copies is quite common. These replicas, however, do not reside at the same locus. As identical in structure as these genes may be, they do not compete with each other in the way that alleles at the same locus can. In the simplest and most basic sense, alleles compete with alternative alleles at the same locus. Any other sorts of competition and cooperation are merely extrapolations from this fundamental sense of allelic competition. Even though genes may cooperate with each other in very complicated ways in embryological development, in replication they can be treated as “separate and distinct.” In development the effects of genes blend. In replication replicators do not blend.
Dawkins introduced the general notions of replicator and vehicle so that selection need not be limited exclusively to gene-based biological evolution. However, as the preceding discussion indicates, his later revisions to his general theoretical outlook were influenced strongly by the traditional perspective of genes and organisms. Genes contain the information necessary to produce organisms and their adaptations. Genes “ride around” in and “guide” organisms. As Dawkins describes them, vehicles are relatively discrete entities that “house” replicators and which can be regarded as machines programmed to preserve and propagate the replicators that ride inside them. Although these terms may be appropriate for the relations between genes and organisms, they interfere with a more general analysis of replication and selection. What really matters in selection is that entities at various levels of organization interact with their environments in such a way that the relevant replicators increase in relative frequency. The actual causal chain that connects replicators and vehicles need not be limited to development.
For example, Dawkins argues at some length that genes and only genes can function as replicators in biological evolution. He goes on to add that “all adaptations are for the preservation of DNA; DNA itself just is” (Dawkins 1982b: 45). But DNA itself exhibits adaptations. Anyone who has spent much time examining the molecular structure of DNA soon realizes that it is adapted to replicate. In addition, the proliferation of junk DNA and meiotic drive are two examples in which the only phenotypes that matter are phenotypic characteristics of genes (Brandon 1996: 133; Sterelny, Smith and Dickison 1996: 388). Dawkins' characterizations of replicators, vehicles and the relations between the two are too closely tied to genes, organisms and development. DNA can certainly replicate itself, but it can also function as a “vehicle” even though it cannot code for, ride around in or direct itself. In sum, a more general characterization of selection is needed, a characterization that does not assume that the only causal connection between replicators and vehicles is development from embryo to maturity. Such a generalization was offered by Hull (1980, 1981, 1988), who proposed that the relevant notion is “interactor” rather than “vehicle”, as interactors are causal and active while vehicles are passive entities.
5. Memes and the Immune System
Initially Dawkins discusses only one example of replication that goes beyond gene-based biological evolution—memetic change. Later he adds the immune system. Does replication play the same role in other sorts of selection that it plays in gene-based biological evolution? The reaction of the immune system to antigens is closest to the standard biological example of replication and selection (Hull 2001). Genes are the primary replicators, but these genes are somatic, not germ-line. Certainly immune systems arose through the same process as other functional systems. The basic structure of the human immune system is “built into our genes,” and the only genes that count in evolution are the ones that we received from our ancestors and can subsequently pass on to our progeny. In Dawkins' terminology, the genes that code for our immune system are active, germ-line replicators.
However, the genes that function in the reaction of the immune system to antigens have two peculiarities. First, they incorporate mechanisms designed to produce very high frequencies of mutation, and second, none of the genes involved in the functioning of the immune system are germ-line. The genes that give rise to B-cells for instance are designed to mutate extensively until one of these cells identifies an invader as foreign. It then proliferates extensively as it attacks the invader. As an organism matures, it accumulates more and more of the B-cells that have been successful in its past. More than this, as the process of proliferation continues, the strength of the affinities to binding sites increases. Initially the primary antibodies almost always exhibit a weak affinity for their targets, but as the reaction to the antigen continues, these affinities become stronger.
Within the confines of a single organism, the reaction of the immune system to antigens has all the characteristics of selection processes, but when the organism dies all of these adaptations are lost. In some species, females pass on not only the genes for the basic structure of their immune systems but also some of the machinery that past invasions of antigens have produced in her. However, these cells are rapidly removed from the offspring as it develops its own immune system. The reaction of the immune system to antigens departs from gene-based selection in biological evolution in two ways. First, from the organismal perspective, the genes that function in protecting an organism from invaders are not germ-line. They are somatic. Second, the relevant mutation rates are much, much higher in the immune system than in ordinary gene-based selection. Instead of mechanisms existing to discover mutations and repair them, mechanisms exist that encourage mutations—massively so. If the functioning of the immune system is to count as selection, then some changes must be made. One possibility is to clarify what counts as a “germ-line” replicator and a rejection of the notion that extremely low mutation rates are inherent in all selection processes.
The modifications needed to include both gene-based selection in ordinary biological evolution and the reaction to the immune system are relatively “minor.” Such is not the case with Dawkins' second example—memetic evolution. Several apparent differences exist between it and the previous examples of replication and selection. Numerous critics have developed a cottage industry engaged in listing all the differences that exist between biological evolution, on the one hand, and social-cultural-conceptual evolution on the other (henceforth, SCC). For example, Dawkins (1976) admits that he is on shaky ground when it comes to the high-fidelity copying required of replicators. Memes seem to get changed much more frequently than genes. However, if differences in copying fidelity are enough to preclude memetic evolution from being a genuine case of selection, then the immune system must also be rejected. Mutation rates are much higher in the production of B-cells than in any area of SCC no matter how speed is calculated.
In general, the standards used to evaluate memetic evolution are much higher than those used to evaluate any other sort of selection. Time and again an overly idealized view of Mendelian genetics is contrasted to a much more realistic view of SCC change. So the story goes, one problem with memes is that they do not have discrete boundaries, do not all come in the same size, and in their functioning are strongly influenced by their environments. Genes, so the critics claim, have sharp boundaries, are all of the same size, and are immune to environmental influences. If memetic evolution is to be evaluated fairly, the same level of criticism must be applied to all putative examples of selection from gene-based selection in biological evolution and the reaction of the immune system to antigens to the development of the central nervous system and social learning (Hull, Langman and Glenn 2001).
Dawkins (1976) also places considerable emphasis on human brains as the “vehicles” for memetic evolution. He defines “meme” as an entity capable of being transmitted from one brain to another. Computers are also plausible vehicles for memes. Dawkins' discussion of memes is, once again, marred by the pervasiveness of the gene-organism perspective. For example, he defines “replicator” in terms of transmission of information—memes leaping from brain to brain or from brains to computers and back again. But memes do not leap from brain to brain or from computer to computer. Their content is transmitted in a variety of ways, including books, audiotapes, conversations and the like. As much as the physical basis changes, the message remains sufficiently unchanged. All instances of this message are equally memes, not just the ones residing in human brains and computers. Memeticists have offered several accounts of memetic replication. Some consider that there is no replication in cultural evolution, but that memes are “attractor points” in culture (Atran 1998) or that cultural transmission is imitation rather than replication (Gatherer 1998). Others consider that the interactor is the meme itself (Blackmore 1999) or that the meme is the selected cultural hereditable information just as Williams' “evolutionary gene” is the selected genetic hereditable information, and the memetic interactor is the repertoire of behaviors it elicits (Wilkins 1998). One view that has been offered several times is that memes are active neural structures (Delius 1991; Aunger 1998, 2002).
All the objections to the gene-meme analogy to one side, Dawkins (1976: 211) finds the chief difference between genetic and memetic change is that biological evolution is at bottom a war between alleles residing at the same locus. “Memes seem to have nothing equivalent to chromosomes and nothing equivalent to alleles.” First, the usual depiction of alleles residing at the same locus on homologous chromosomes so central to Mendelian genetics is an oversimplification, but more importantly, for at least half of life on earth, replication and selection took place in the absence of chromosomes, meiosis and the like. If gene-based biological evolution took place for so long in the absence of the Mendelian apparatus and still does so in many extant organisms, then just possibly we should not demand that memetic evolution proceed by this very special and possibly aberrant sort of inheritance. The cost of meiosis remains a serious problem in ordinary biological evolution. Demanding that SCC evolution incorporate this same highly problematic element in its own makeup seems strange in the extreme. If we are to develop a general analysis of selection, then we must distinguish between essential and contingent features of this process.
Numerous evolutionary biologists question how fundamental to selection the perspective of alleles at a locus actually is. Almost everyone agrees that evolution involves changes in gene frequencies. However, few go on to add that evolution is nothing but changes in gene frequencies. When one looks at the work of evolutionary biologists, one discovers that it involves much more than changes in gene frequencies. Selection in meme-based evolution must be fleshed out. It remains to be seen how different well-worked-out versions of SCC theory will differ from more familiar forms of selection. And where they differ, it does not follow automatically that the meme-based theory must be modified. One might well change the traditional gene selectionist view of biological evolution. Selection in gene-based theories of evolution was worked out first, but historical precedence does not entail conceptual priority.
6. The Extended Replicator
The discussion thus far has involved criticisms of Dawkins' notion of genes and replication. Dawkins' critics think that genes play too important of a role in his notion of replication. Replication, so they argue, can occur at other levels of organization as well. Just as Dawkins extended the notion of the phenotype, these authors propose to make the notion of replicators more general as well—to extend the replicator so to speak. Dawkins introduced his Extended Phenotype conception for two reasons. First, he wanted to extend the notion of a “phenotypic trait.” The sort of nest that a bird builds or the song that it sings can count as phenotypic traits just as much as the shape of its bill. Second, Dawkins wanted to break the hold that organisms have over how we conceptualize the living world. Traits do tend to come bundled into reasonably discrete entities, but for making inferences about the evolutionary process, traits can be treated as separate and distinct. However, as he proceeded, Dawkins eventually decided that extending the phenotype was not enough. He had to bury it. All sorts of fascinating mechanisms to one side, what really matters in selection takes place at the level of replicators.
Although such critics of Dawkins as Sterelny et al. (1996) decline to join with Dawkins in his demotion of environmental interaction, they do agree with him that replicators are special. They play a special role in development. However, they do not limit replicators to genes even in biological evolution. Sterelny et al. (1996) propose to extend the replicator to include nonstandard entities: “the set of developmental resources that are adapted from the transmission of similarity across the generations” (Sterelny 2001a: 338). For example, the sort of burrow that a particular organism digs is influenced by its genetic makeup, but if these burrows are used over and over again, characteristics of these burrows can themselves be viewed as replicators. The effects of these burrows get passed on from generation to generation, but not via the genes.
As more and more nongenetic replicators, including epigenetic inheritance, are acknowledged, Dawkins' Gene's Eye view begins to gradate into the conception of the Extended Replicator, which Sterelny et al. (1996) specify as any non-genetic as well as genetic replicators. These two views are so general that any case that can be described in one can be redescribed in the other. Differences lie in ease of description. According to Sterelny et al. (1996), the burrows that some organisms dig can function as replicators—extended replicators—while Dawkins portrays them as instance of extended phenotypes. The contrast is between selfish burrows and selfish genes for burrowing.
In the wake of Oyama's The Ontogeny of Information (1985) and earlier work, notably that of Daniel Lehrmann (2001) and Patrick Bateson (2001), a view of biological evolution has arisen that emphasizes development (see also Griffiths and Gray 1994; Oyama 2000). In the 19th century development was an extremely active research program. The next great discoveries in biology were going to be in the area of development. It was not to be. First Mendelian genetics and then a version of evolutionary theory centered on population genetics took over biology, and they did so while avoiding development. Everyone knew that development was central to both evolution and reproduction, but no one could see how to integrate the masses of developmental data available into the emerging synthesis of evolutionary biology and genetics. As a result, development was left out of the New Synthesis (Buerton et al. 2000; Gilbert 1991). Considering how central development actually is in biology, the advances made while ignoring it are staggering.
Even so, developmental biology continued on its course until at long last we seem to understand development well enough to begin integrating it into the rest of biology. On the most conservative view, current versions of evolutionary theory can remain largely unchanged as development is grafted onto them. On a second view, both perspectives are likely to require some modification to bring off this integration. Our understanding of development may have to be modified, but so too for evolutionary theory (Pigliucci and Muller 2010). Finally, at the other extreme, development will all but replace evolutionary theory. In their more exuberant moments, some advocates of Developmental Systems Theory (hereafter DST) seem to be claiming just that. Just as some molecular biologists think that molecular biology is rapidly replacing all the rest of biology, some advocates of DST argue that developmental theories will simply replace current versions of evolutionary theory.
On some versions of the DST view, genes have no uniquely privileged role in repeated cycles of development (Griffiths and Gray 1994; Griffiths 2001). In fact, no element of the developmental matrix plays any privileged causal role—not genes, not organisms, not the environment, not anything. Everything counts as a resource, albeit in particular situations certain resources will play more important roles than other resources. In rejecting any privileged role for genes, advocates of DST are especially skeptical of one particular role supposedly played by genes—the transmission of information. According to some, information is central to developmentalism, but genes are not the only mechanisms for information transfer. According to others, information plays no role in the emerging developmentalist perspective. The developmental system as a whole is the unit of selection.
In the continuing debate over developmental versus traditional theories of evolutionary biology, Sterelny et al. (1996) hold a fairly conservative position. They agree with the developmentalists that genes play no privileged role in the development of phenotypes from genotypes. Genes play a role in this process, simply not a privileged role. Genes can serve as a causal bridge from phenotype to phenotype, but other entities can do so as well. Genes are not the only replicators in biological evolution. The repeated cycles in inheritance include many different sorts of constancies and repetitions—genes, cellular machinery, phenotypic traits including behaviors, and social structures. Information remains central to selection processes, but genes are not the only carriers of such information. Genes predict phenotypic characters only in the same sense that environmental factors predict them. More recently, Sterelny (2000b) has argued that developmental considerations require no fundamental reevaluation of evolutionary conceptions along the lines of DST.
Both Dawkins (1976) and Hull (1980) distinguish between replicators and vehicles (or interactors for Hull). The distinction, now referred to as the Hull-Dawkins Distinction, has broad acceptance across evolutionary debates (Eldredge 1989), although DST challenges its necessity for all cases, and its primacy in conceiving evolutionary questions. To the questions surrounding these two sorts of entities, Lloyd (1992) adds two more—who “owns” adaptations and who “benefits” from selection? In the previous literature, authors seem to assume a greater concordance in the answers to these four questions than can be justified. Some biologists argue that those entities that exhibit a particular adaptation are the beneficiaries of the selection process that resulted in these adaptations. Dawkins disagrees. Such adaptations function only as intermediaries in a much more fundamental process—the spreading of copies of particular alleles. These surviving alleles are the long-term beneficiaries of selection processes. Sterelny et al. (1996) argue that replicators and only replicators are the beneficiaries of adaptations, keeping in mind that they count entities more inclusive than genes as replicators. Gould, following Wimsatt, even goes so far as to argue that the causal agents in selection are the interactors, and that genes and other replicators are “important in evolution, but in a different role as items for bookkeeping. Replicators are not causal agents” (2002: 616, 632n gives his link to Wimsatt. See Wimsatt and Griesemer 2007). Of course, replicators must play some causal role, unless Gould is supposing that as replicators they are merely conceptual entities, but they do not, he thinks, play a role as units of selection.
With respect to the version of replication formulated by Sterelny et al. (1996), both copying and biofunction are crucial. Copying is quite obviously a causal phenomenon, but not any old causal connection will do. Similarity of copies is necessary, but not sufficient. These copies must be copies of copies. One copy must produce another copy and that copy produce still another copy and so on. For one entity to be a copy of another, it must be the output of a process whose biofunction is to conserve function. The function of copying is to produce from one token another token which is similar in the relevant respects. Genes fit this definition, but so do lots of examples of nongenetic transmission; e.g., habitat stability resulting from nest site imprinting, the song that a bird learns, various micro-organism symbionts, not to mention SCC transmission.
7. Information
The language of “codes” and “information” flows easily enough with respect to replication. Transcription, translation, punctuation, redundancy, synonymy, editing, proofreading, errors, repairing of errors, messages, copies, and information all sound natural enough. This ease of expression supports the contention that selection plays the same role in SCC evolution as it does in gene-based biological evolution. Some authors argue that both processes count as languages in the same sense of “language.” All the problems listed for treating gene-based biological evolution as a language are simply replicated in ordinary languages. If the genetic code is not a code, then neither is the Morse code or English for that matter. Differences of degree exist, but not differences in kind. For example, mutation rates are very low in gene-based biological evolution, moderate in SCC evolution and massive with respect to the immune system. Concurrent variation within loci is extensive in the immune system, greatly reduced in both gene-based biological evolution and SCC evolution, and reduced even further in operant learning. Others deny that DNA presents some relevant properties of language, such as Zipf's law (Tsonis, Elsner, and Tsonis 1997, see also entry on biological information).
The literature dealing with information is both extensive and factious. Several different formal analyses of information can be found and very little agreement about which analysis is best for which subjects. On one point these scholars tend to agree—cybernetic information and communication-theoretic information will not do for replication in biological contexts. The best bet is semantic information (Sterelny 2000a; Godfrey-Smith 2000; Sarkar 2000). The trouble is that no widely accepted version of semantic information exists. Winnie (2000) distinguishes between Classical and Algorithmic Information Theory and opts for a revised version of the Algorithmic Theory. But once again, the problem is that no such formal analysis currently exists. In the face of all this disagreement and unfinished business, biologists such as Maynard Smith (2000) maintain either that informal analyses of “information” are good enough or that some future formal version of information theory will justify the sorts of inferences that they make. The sense of “information” as used in the Central Dogma of molecular biology, which states that information cannot flow from protein to DNA, is more like a fit of template, or the primary structure of the protein sequence compared to the sequence of the DNA base pairs. Attempts have been made in what is now known as bioinformatics to use Classical Information Theory (Shannon's theory of communication) to extract functional and phylogenetic information (Gatlin 1972; Maclaurin 1998; Wallace and Wallace 1998; Brooks and Wiley 1988), but it appears to have been unsuccessful in the main. While the most likely conclusion is that no version of information theory as currently formulated can handle “information” as it functions in biology (see Griffiths 2001 for further discussion), attempts have been made to formulate just such a version (Sternberg 2008; Bergstrom and Rosvall 2011). However, this undercuts the motivation for appealing to information theory to elucidate genes in the first instance.
8. Reproducers
In recent developments, the notion of a reproducer has been proposed by James Griesemer (Griesemer 2000a,b, 2002, 2005; Wimsatt and Griesemer 2007). A reproducer is an entity that develops and has a material overlap between the “parent” and “progeny”. Maynard Smith and Szathmáry first published a definition of reproducer, based on Griesemer's definition (Szathmáry and Maynard Smith 1997):
- x begets y iff y ≠ x and x is (part of) the efficient cause of y (where x and y always refer to individuals).
- x materially overlaps y iff there is a physical part z, of x at time t which is a physical part of y at time t ≠ t′.
- x progenerates y iff x begets y and x materially overlaps y.
- Reproduction is a process of progeneration the result of which is an increase in numerically distinct objects of a given kind.
- x is a reproducer iff x was the product of progeneration and x has the capacity to develop the capacity to progenerate.
- Reproduction is a composite process of progeneration and development, the result of which is an increase in numerically distinct objects with the capacity to develop the capacity to progenerate (or progenerate directly).
- x is a reproducer iff x was the product of progeneration and x has the capacity to reproduce.
- x acquires something, p (a part, a property), iff p is not a part/property of x at time t, but p is part/property of x at some later time t′.
- x develops iff x acquires the capacity to reproduce.
Here inheritance is a system property, not a property of parts unless they are reproducing systems, and is developmentally acquired. Organisms do not arise already capable of reproduction in the main, but must undergo orderly transitions before they may reproduce.
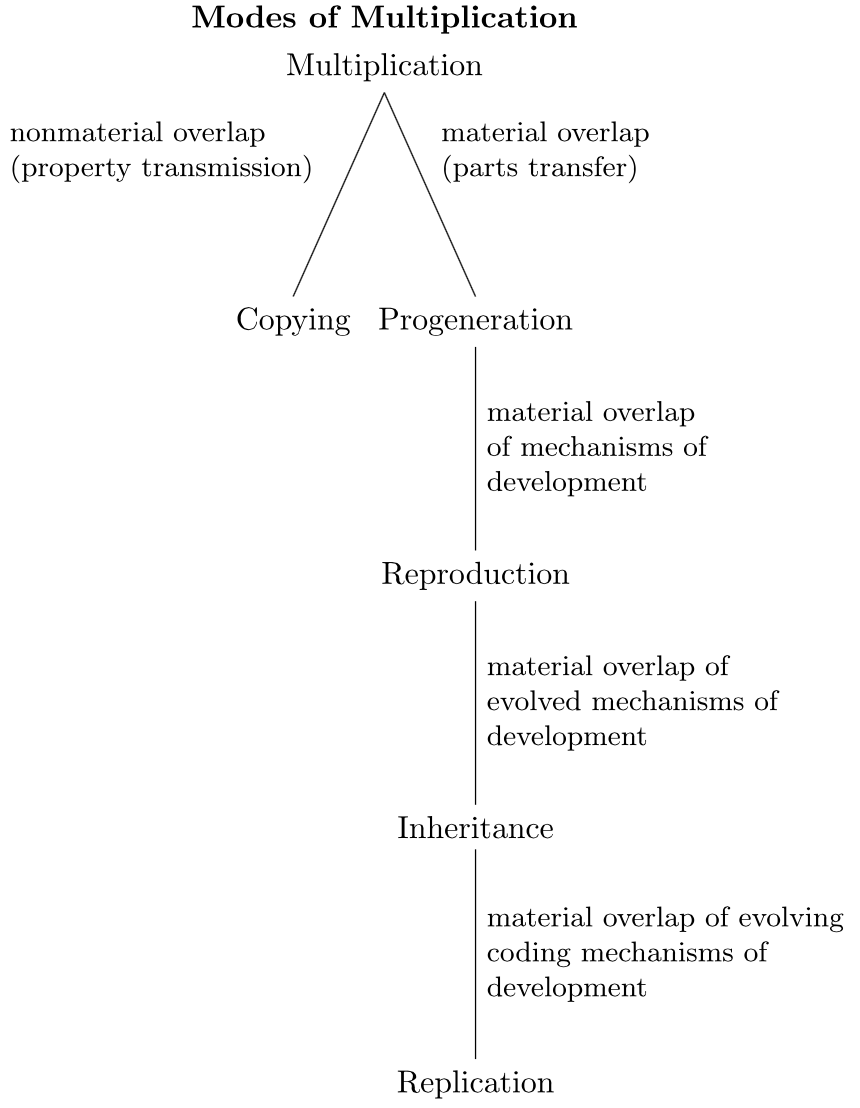
The relation between reproducers, and replicators and interactors, varies according to author. Griesemer treats replication as the terminal form of reproduction, once coding mechanisms have been evolved, which is itself an evolved form of multiplication (Griesemer 2000b: 76). Unlike Dawkins' replicator concept, which Griesemer believes is based on a similarity relationship of “copying”, a reproducer requires material overlap of systems. However, replication arises when material overlap evolves progeneration, and progeneration evolves a codical inheritance system, as the above figure (from Wimsatt and Griesemer 2007: 268) indicates.
Others, particularly Jablonka and Lamb (2005, 2007), think that reproducers are distinct from replication, and that some epigenetic inheritance systems are reproducers without being replicators. For them, a reproducer is the “target of selection”.
Godfrey-Smith (2009: 83) takes issue with Griesemer's requirement of “material overlap” as being too narrow a definiendum of reproducers. He thinks that reproducers need not contribute actual material to their progeny (as in cases where they template for sequences in the case of viruses). Griesemer believes Godfrey-Smith has over-interpreted the requirement; he is primarily concerned with the idea that parents cause progeny (Griesemer 2014).
An aspect of reproducers that has fascinated thinkers is that it does not imply a particular level of reproduction or progeneration, and indeed Szathmáry and Maynard Smith consider that the major transitions of evolution each involve an evolved mode of progeneration that is more inclusive or novel than what it evolved from (Szathmáry and Maynard Smith 1997; Maynard Smith and Szathmáry 1998). The notion of reproducers has also been applied to speciation and cladogenesis (Hamilton and Haber 2006). While it may apply to populations, it is unlikely to apply to species or supraspecific taxa, and macroevolution in general.
9. Conclusion
In general the authors who have concerned themselves with the role of replication in selection strive to liberate our thinking from the hold that genes and organisms have over us. To the extent that replication can be separated off from development, it plays a central role in selection processes of all sorts, including traditional gene-based biological evolution, the reaction of the immune system to antigens and possibly even SCC evolution. In reaction to all sorts of nonstandard examples of replication, Dawkins is driven to extend the phenotype. In reaction to DST and evolutionary developmental biology some critics of Dawkins extend the replicator as well. Replicators include genes, memes and even entities that are commonly thought of as parts of an organism's phenotype or environment. Some authors argue that replication in this extended sense is necessary for selection; others that it is not. Selection can occur in the absence of replication. Still others maintain that all of these biological phenomena are best explained without any reference to such traditional notions as replication, environmental interaction, selection, evolution, etc., in terms of DST. Others, using the notion of reproducers, treat replication as an embodied process of coding.
Bibliography
- Atran, S., 1998, “Folk Biology and the Anthropology of Science: Cognitive Universals and the Cultural Particulars”, Behavioral and Brain Sciences, 21(4): 547–609.
- Aunger, R., 1998, “The ‘Core Meme’ Meme”, Behavioral and Brain Sciences, 21(4): 569–570.
- –––, 2002, The Electric Meme: A New Theory of How We Think, New York: Free Press.
- Bateson, P., 2001, “Behavioral Development and Darwinian Evolution”, in Oyama et al. 2001, pp. 149-166.
- Bergstrom, C. and M. Rosvall, 2011, “The transmission sense of information”, Biology and Philosophy, 26(2): 159–176.
- Blackmore, S. J., 1999, The Meme Machine, Oxford; New York: Oxford University Press.
- Brooks, D. R. and E. O. Wiley, 1988, Evolution as Entropy: Toward a Unified Theory of Biology, edited by D. L. Hull, 2nd edition (Science and its Conceptual Foundations), Chicago: University of Chicago Press.
- Brandon, R. R., 1996, Concepts and Methods in Evolutionary Biology, Cambridge: Cambridge University Press.
- Beurton, Peter, Raphael Falk, and Hans-Jörg Rheinberger, 2000, The concept of the gene in development and evolution: historical and epistemological perspectives, (Cambridge studies in philosophy and biology), Cambridge: Cambridge University Press.
- Caporael, L., 2003, “Repeated Assembly,” in Evolutionary Psychology, edited by S.J. Scher and F. Rauscher, New York: Springer US, pp. 71–89.
- Cook, P. R., 1999, “The Organization of Replication and Transcription”, Science, 284: 1790–1797.
- Crick, F., 1970, “Central dogma of molecular biology”, Nature, 227(5258): 561–563.
- Dawkins, R., 1976, The Selfish Gene, Oxford: Oxford University Press.
- –––, 1982a, The Extended Phenotype, Oxford: Oxford University Press.
- –––, 1982b, “Replicators and Vehicles”, in Current Problems in Sociobiology, edited by King's College Sociobiology Group, Cambridge: Cambridge University Press, pp. 45–64.
- –––, 1994, “Burying the Vehicle”, Behavioral and Brain Sciences, 17: 616–617.
- –––, 1999, The Selfish Gene, Oxford: Oxford University Press, Revised edition. Original edition in 1976.
- Delius, J., 1991, “The Nature of Culture”, in The Tinbergen Legacy, edited by M. S. Dawkins, T. R. Halliday and R. Dawkins, London: Chapman and Hall.
- Eldredge, N., 1989, Macroevolutionary Dynamics: Species, Niches, and Adaptive Peaks, New York: McGraw-Hill.
- Fisher, R. A., 1930, The Genetical Theory of Natural Selection, Oxford UK: Clarendon Press, (rev. ed. Dover, New York, 1958).
- Gatherer, D., 1998, “Why the ‘Thought Contagion’ Metaphor is Retarding the Progress of Memetics”, Journal of Memetics—Evolutionary Models of Information Transmission, 2. [Gatherer 1998 available online]
- Gatlin, L. L., 1972, Information Theory and the Living System, New York: Columbia University Press.
- Gilbert, Scott F., 1991, A Conceptual history of modern embryology, Baltimore: Johns Hopkins University Press.
- Godfrey Smith, P., 2000, “Information, Arbitrariness, and Information: Comments on Maynard Smith”, Philosophy of Science, 67: 202–207.
- –––, 2009, Darwinian populations and natural selection, Oxford: Oxford University Press.
- Godfrey-Smith, P. and B. Kerr, 2002, “Individualist and Multi-level Perspectives on Selection in Structured Populations”, Biology and Philosophy, 17(4): 477–517, with commentaries, and replies by the authors, pp. 539–550.
- Gould, S. J., 2002, The Structure of Evolutionary Theory, Cambridge, MA: Harvard University Press.
- Griesemer, J. R., 2000a, “Development, Culture, and the Units of Inheritance”, Philosophy of Science, 67 (Supplement. Proceedings of the 1998 Biennial Meetings of the Philosophy of Science Association. Part II: Symposia Papers): S348–S368.
- –––, 2000b, “The units of evolutionary transition”, Selection, 1(1–3): 67–80.
- –––, 2002, “What Is ‘Epi’ about Epigenetics?” Annals of the New York Academy of Sciences, 981(1): 97–110.
- –––, 2005, “The informational gene and the substantial body: on the generalization of evolutionary theory by abstraction”, in Idealization XII: Correcting the Model. Idealization and Abstraction in the Sciences, edited by M. R. Jones and N. Cartwright. Amsterdam: Rodopi Publishers: 59–115.
- –––, 2014, “Reproduction and the Scaffolded Development of Hybrids”, in Developing Scaffolds in Evolution, Culture, and Cognition, edited by Linnda Caporael, James R. Griesemer and William C. Wimsatt, Cambridge, MA: MIT Press.
- Griffiths, P., 2001, “Genetic Information: A Metaphor in Search of a Theory”, Philosophy of Science, 68: 394–412.
- Griffiths, P. and R. Gray, 1994, “Developmental Systems and Evolutionary Explanation”, Journal of Philosophy, 91: 277–304.
- Hamilton, W. D., 1964, “The genetical evolution of social behaviour. I”, Journal of Theoretical Biology, 7(1): 1–16.
- Hamilton, A. and M. H. Haber, 2006, “Clades Are Reproducers”, Biological Theory, 1(4): 381–391.
- Hodge, M. J. S., 1989, “Generation and the Origin of Species (1837–1937): A Historiographical Suggestion”, British Journal for the History of Science, 22(74): 267–281.
- Hull, D. L., 1980, “Individuality and Selection”, Annual Review of Ecology and Systematics, 11: 311–332.
- –––, 1981, “Units of evolution: a metaphysical essay”, in The philosophy of evolution, edited by U.L. Jensen and R. Harré, Brighton UK: Harvester Press, pp. 23–44.
- –––, 1988, “Interactors versus vehicles”, in The role of behavior in evolution, edited by H. C. Plotkin, Cambridge, MA: MIT Press, pp. 19–50.
- –––, 2001, Science and selection: Essays on Biological Evolution and the Philosophy of Science, (Cambridge studies in philosophy and biology), Cambridge, UK; New York: Cambridge University Press.
- Hull, D. L., R. E. Langman, and S. S. Glenn, 2001, “A General Account of Selection: Biology, Immunology, and Behavior”, Behavioral and Brain Sciences, 24: 511–573.
- Jablonka, E. and M. J. Lamb, 2005, Evolution in four dimensions: genetic, epigenetic, behavioral, and symbolic variation in the history of life, Life and mind, Cambridge, MA: MIT Press.
- –––, 2007, “Précis of Evolution in Four Dimensions”, Behavioral and Brain Sciences, 30(4): 353–365.
- Jablonka, E. and G. Raz, 2009, “Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution”, The Quarterly Review of Biology, 84(2): 131–76.
- Jacob, F., 1973, The logic of life: a history of heredity, New York: Pantheon Books.
- Jacobson, H., 1958, “On models of reproduction”, American Scientist, 46(3): 255–284.
- Keller. L. (ed.), 1999, Levels of Selection, Princeton: Princeton University Press.
- Kerr, B. and P. Godfrey-Smith, 2002, “Individualist and Multi-level Perspectives on Selection in Structured Populations”, Biology and Philosophy, 17: 477–517.
- Lehrman, D. S., 2001, “A Critique of Konrad Lorenz's Theory of Instinctive Behavior”, in Oyama et al. 2001, pp. 25-40.
- Lewes, G. H., [Anon.], 1856, “Hereditary Influence, Animal and Human”, Westminster Review, 66(July): 135–162.
- Lloyd, E. A., 1992, “Unit of Selection”, in Keywords in Evolutionary Biology, edited by E. Fox Keller and E. A. Lloyd, Cambridge, MA: Harvard University Press, pp. 334–340.
- –––, 1994 [1988], The structure and confirmation of evolutionary theory, Princeton, NJ: Princeton University Press. Original edition 1988, New York: Greenwood Press.
- Maynard Smith, J., 2000, “The Concept of Information in Biology”, Philosophy of Science, 67: 177–194.
- Maynard Smith, J. and E. Szathmáry, 1998, The origins of life: from the birth of life to the origins of language, Oxford: Oxford University Press.
- Maclaurin, J., 1998, “Reinventing Molecular Weismannism: Information in Evolution”, Biology and Philosophy, 13(1): 37–59.
- Mitchell, S.D., 2009, Unsimple truths: science, complexity, and policy, Chicago: University of Chicago Press.
- Okasha, S., 2004, “The ‘Averaging Fallacy’ and the Levels of Selection”, Biology and Philosophy, 19: 167–184.
- –––, 2005, “Multilevel Selection and the Major Transitions in Evolution”, Philosophy of Science, 72(5): 1013–1025,
- –––, 2006, Evolution and the levels of selection, Oxford: Clarendon.
- Oyama, S., 1985, The Ontogeny of Information: Developmental Systems and Evolution, Cambridge: Cambridge University Press.
- –––, 2000, The Ontogeny of Information: Developmental Systems and Evolution, Second Edition, revised and expanded with an Introduction by Richard C. Lewontin, Durham, North Carolina: Duke University Press.
- Oyama, S., P. E. Griffiths, and R. D. Gray (eds.), 2001, Cycles of contingency: Developmental systems and evolution, Cambridge, MA: MIT Press.
- Pigliucci, M. and G. Muller, 2010, Evolution: The Extended Synthesis, Cambridge, MA: MIT Press.
- Rebek, Jr., J., 1994, “Synthetic Self-Replicating Molecules”, Scientific American, 271: 48–55.
- Sarkar, S., 2000, “Information in Genetics and Developmental Biology: Comments on Maynard Smith”, Philosophy of Science, 67: 208–213.
- Schrödinger, E., 1969 [1944], What is life? The physical aspect of the living cell, with Mind and matter, Cambridge UK: Cambridge University Press. Original edition, 1944.
- Shannon, C. E., 1948, “A mathematical theory of communication”, The Bell System Technical Journal, 27: 379–423, 623–656.
- Sterelny, K., 2000a, “The ‘Genetic Program’: A commentary on Maynard Smith on Information in Biology”, Philosophy of Science, 67: 195–201.
- –––, 2000b, “Development, evolution, and adaptation”, Philosophy of Science, 67(Supplement. Proceedings of the 1998 Biennial Meetings of the Philosophy of Science Association): S369–S387.
- –––, 2001a, The Evolution of Agency and Other Essays, Cambridge: Cambridge University Press.
- –––, 2001b, “Niche Construction and the Extended Replicator”, in Oyama, Griffiths, and Gray 2001, pp. 333–349.
- Sterelny, K., K. C. Smith and M. Dickison, 1996, “The Extended Replicator”, Biology and Philosophy, 11: 377–403.
- Sternberg, R., 2008, “DNA Codes and Information: Formal Structures and Relational Causes”, Acta Biotheoretica, 56(3): 205–232.
- Šustar, P., 2007, “Crick's notion of genetic information and the ‘central dogma’ of molecular biology”, British Journal for the Philosophy of Science, 58(1): 13–24.
- Szathmáry, E. and J. Maynard Smith, 1997, “From Replicators to Reproducers: the First Major Transitions Leading to Life” Journal of Theoretical Biology, 187(4): 555–571.
- Tsonis, A. A., J. B. Elsner, and P. A. Tsonis, 1997, “Is DNA a Language?”, Journal of Theoretical Biology, 184: 25–29.
- Van Regenmortel, M. and D. L. Hull, 2002, Promises and Limits of Reductionism in the Biomedical Sciences, Chichester: John Wiley and Sons.
- von Neumann, J. and A. W. Burks, 1966, Theory of self-reproducing automata, edited and completed by Arthur W. Burks, Urbana, London: Illinois University Press.
- Wallace, R. and R. G. Wallace, 1998, “Information Theory, Scaling Laws and the Thermodynamics of Evolution”, Journal of Theoretical Biology, 192: 545–559.
- Wiener, N., 1948, Cybernetics, or, Control and communication in the animal and the machine, Cambridge, MA: Technology Press.
- Wilkins, J. S., 1998, “What's in a Meme? Reflections from the Perspective of the History and Philosophy of Evolutionary Biology”, Journal of Memetics—Evolutionary Models of Information Transmission, 2. [Wilkins 1998 available online]
- Williams, G. C., 1966, Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought, Princeton, NJ: Princeton University Press.
- Wimsatt, W. C. and J. R. Griesemer, 2007, “Reproducing Entrenchments to Scaffold Culture: The Central Role of Development in Cultural Evolution”, in Integrating Evolution and Development: From Theory to Practice, edited by R. Sansom and R. Brandon, Cambridge: MIT Press: 227–323.
- Winnie, J. A., 2000, “Information and Structure in Molecular Biology: Comments on Maynard Smith”, Philosophy of Science, 67: 517–526.
Academic Tools
How to cite this entry. Preview the PDF version of this entry at the Friends of the SEP Society. Look up this entry topic at the Indiana Philosophy Ontology Project (InPhO). Enhanced bibliography for this entry at PhilPapers, with links to its database.
Other Internet Resources
- The Artificial Self-Replication page, maintained by Dr. Moshe Sipper (Logic Systems Lab, Swiss Federal Institute of Technology). The concept of replication has led a largely independent life in the formal sciences as a consequence of John von Neumann's seminal work on self-replicating automata. This page offers a good introduction to the literature.
- A good introduction to the molecular aspects of DNA replication is atdbio's.
Acknowledgments
The original version of this article, entitled “Replication” was written by the late David Hull. In January 2005, John Wilkins became a co-author, with the responsibility for revising and updating the entry, so as to keep it current. Thus, revisions on or after January 26, 2005 were introduced by Wilkins. The current version represents a significant revision of Hull's original essay and may not represent his final views.
The editors would like to thank Christopher von Bülow for reading this entry carefully and pointing out a number of typographical errors. The author would like to thank Peter Godfrey-Smith and James Griesemer for their suggestions and comments, and the reviewer who made many useful comments and suggestions.